Fascinating! Just ordered 3 tests. My newest tank had a brief bout of dinos while the other two are stable although some corals never seem to do well in one of them. It will be very interesting comparing the results.
Navigation
Install the app
How to install the app on iOS
Follow along with the video below to see how to install our site as a web app on your home screen.
Note: This feature may not be available in some browsers.
More options
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
This is what I've dreamed of for so long! Testing for microbes in our tanks!
- Thread starter Flippers4pups
- Start date
- Tagged users None
Flippers4pups
Fins up since 1993
View BadgesExcellence Award
Reef Tank 365
Article Contributor
Reef Squad Emeritus
Hospitality Award
Hi everyone,
As the owner of the company I'd be happy to answer any questions. There have been a lot asked already in the thread so forgive me if I miss some. I'd like to start with a few of the more important points.
1. I promise: no snake oil. I'm selling a testing service; whoever compared it to ICP is exactly right.
You will notice my site is conspicuously absent of promises that "this bacterium is bad" or "this bacterium is good". We simply don't know enough yet to make those claims.
2. By building a large database of samples from hobbyist and aquaculture tanks, we will learn a lot. I already have some data in hand and am gathering more.
3. There is some skepticism about what this means. Skepticism is a wise default position, but please do note what my service claims or does not claim. I claim to be able to identify the microbes in your tank, and provide information about what those microbes do. I will stand by those claims, since they are the basis for most modern microbial ecology.
I do NOT claim that I can sell you a magic bottle that will make everything happy. Or even that we understand everything about the microbes we find. In the reports, I try hard to not overstate the conclusions.
I've spent the last 20 years in Academia, so I'm comfortable with disagreement and discussion. Don't be shy about disagreeing - my service is simply DNA sequencing, I'm not selling bacteria. So I won't be offended if someone has an argument why bacterial type X is unimportant. (If there is someone who thinks it doesn't matter what kind of microbes are in your tank, I would disagree, but I'm not sure anyone in the reefing world holds that opinion anyway! And it would be an interesting discussion in any case)
4. Finally (for this post), some have raised the question of planktonic versus surface associated bacteria. Its an important distinction. But please recognize that water is constantly circulating past the surfaces in our tanks -- the biofilm microbes show up in the water column too. Lots of evidence in hand now to show this.
With that said, our first round did show that for a more sensitive analysis of biofilm microbes, we should include a direct sample of the biofilm. This contains important ammonia-oxidizing and nitrate-oxidizing microbes. So current sampling kits include a biofilm sample too.
I'm gonna stop there for now, but I'll come back and answer questions. There is a link on the main page of the company website that describes a basic overview of the microbes that live in reef tanks. Soon I'll be adding my analysis of differences between tanks, and my cycling experiment studying the succession of microbes in new aquariums started various ways.
We have a lot to learn - but the way we learn is by collecting data. Thats what my service is for.
-Eli
Thank you for your reply and straight forward answers! And for joining us here! You are always welcome!
- Joined
- Oct 3, 2015
- Messages
- 5,145
- Reaction score
- 8,758
Hi everyone,
As the owner of the company I'd be happy to answer any questions. There have been a lot asked already in the thread so forgive me if I miss some. I'd like to start with a few of the more important points.
1. I promise: no snake oil. I'm selling a testing service; whoever compared it to ICP is exactly right.
You will notice my site is conspicuously absent of promises that "this bacterium is bad" or "this bacterium is good". We simply don't know enough yet to make those claims.
2. By building a large database of samples from hobbyist and aquaculture tanks, we will learn a lot. I already have some data in hand and am gathering more.
3. There is some skepticism about what this means. Skepticism is a wise default position, but please do note what my service claims or does not claim. I claim to be able to identify the microbes in your tank, and provide information about what those microbes do. I will stand by those claims, since they are the basis for most modern microbial ecology.
I do NOT claim that I can sell you a magic bottle that will make everything happy. Or even that we understand everything about the microbes we find. In the reports, I try hard to not overstate the conclusions.
I've spent the last 20 years in Academia, so I'm comfortable with disagreement and discussion. Don't be shy about disagreeing - my service is simply DNA sequencing, I'm not selling bacteria. So I won't be offended if someone has an argument why bacterial type X is unimportant. (If there is someone who thinks it doesn't matter what kind of microbes are in your tank, I would disagree, but I'm not sure anyone in the reefing world holds that opinion anyway! And it would be an interesting discussion in any case)
4. Finally (for this post), some have raised the question of planktonic versus surface associated bacteria. Its an important distinction. But please recognize that water is constantly circulating past the surfaces in our tanks -- the biofilm microbes show up in the water column too. Lots of evidence in hand now to show this.
With that said, our first round did show that for a more sensitive analysis of biofilm microbes, we should include a direct sample of the biofilm. This contains important ammonia-oxidizing and nitrate-oxidizing microbes. So current sampling kits include a biofilm sample too.
I'm gonna stop there for now, but I'll come back and answer questions. There is a link on the main page of the company website that describes a basic overview of the microbes that live in reef tanks. Soon I'll be adding my analysis of differences between tanks, and my cycling experiment studying the succession of microbes in new aquariums started various ways.
We have a lot to learn - but the way we learn is by collecting data. Thats what my service is for.
-Eli
Welcome to R2R! What a great response. Look forward to hearing more about your findings.
- Joined
- Sep 21, 2018
- Messages
- 6,657
- Reaction score
- 7,143
I think the obvious huge limitation is that even if one could somehow get a numerical analysis of all planktonic species, it is believed that the majority of bacteria are attached to surfaces of various types.
What about DNA from the bacteria on the surfaces getting into the water column? Does the analysis somehow only pickup DNA from suspended organisms?
I found this exert from Eli's analysis very interesting
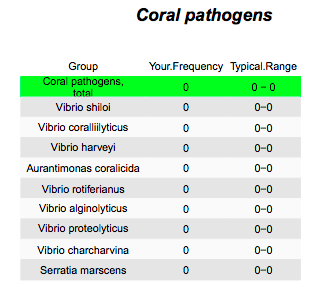
"We screened for 9 different coral pathogens, and found no evidence of these in any tanks sampled for this study. "
"We screened for 9 different coral pathogens, and found no evidence of these in any tanks sampled for this study. "
Although admittedly conjecture (on my part anyway), I would also presume that most microbes present on surfaces will have a smaller presence in the water column. Exceptions might be anaerobes buried deep in rocks or sediment. However, even if present in the water column, if the numbers are small enough it's possible that a single water sample could contain too few organisms for detection.
In any case, this test also includes sampling from a surface location in your tank.
One thing that will be interesting is to see the relative abundance of vibrio and mycobacterial DNA in the samples; two groups that can potentially infect the aquarist.
In any case, this test also includes sampling from a surface location in your tank.
One thing that will be interesting is to see the relative abundance of vibrio and mycobacterial DNA in the samples; two groups that can potentially infect the aquarist.
Randy Holmes-Farley
Reef Chemist
View Badges
Staff member
Super Moderator
Excellence Award
Expert Contributor
Article Contributor
R2R Research
My Tank Thread
- Joined
- Sep 5, 2014
- Messages
- 67,160
- Reaction score
- 63,517
Not true. There is also a sample swabbed from the output pipe of sampled tanks. He added this for the latest sampling method.
It should also be noted that in re-circulating saltwater systems, pretty much all bacteria can be sampled from the water column, not just planktonic bacteria.
He is confirming this with swab tests.
EDIT: I see Eli just posted something similar as I was typing.
That’s a fine assertion. How do you know it is true?
Different environments will also have different species. The surface of a pipe will be very different than down in sand, on an organism like macroalgae, in light vs dark, etc.
I participated in his early rounds of testing, as he's a member of my local aquarium club. Very nice guy, and I don't think he's into offering any "snake oil" remedies, and he's definitely scientific in his approach on testing and comparing tanks. Here are some snippets from the report on my tank, just to give you an idea of what to expect:
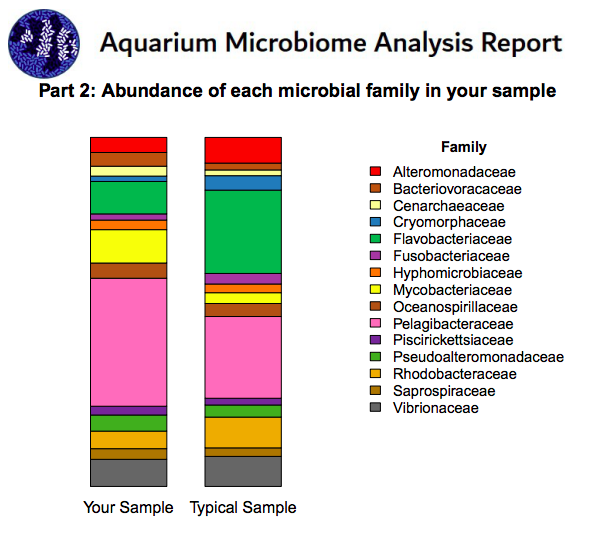
Dude.. you know you are going to seed my rock for the new tank right
- Joined
- Oct 3, 2015
- Messages
- 5,145
- Reaction score
- 8,758
So are we saying that we should all share little vials of tank water with each other, to share microbes and have maximum variation?
I could see a little "Vial swap" stand setup at frag swaps lol
I could see a little "Vial swap" stand setup at frag swaps lol
SuncrestReef
That Apex guy
View BadgesExcellence Award
Reef Tank 365
Article Contributor
Reef Squad Emeritus
Controller Advisor
Dude.. you know you are going to seed my rock for the new tank right
Rumor is that I have the most biodiverse rock in the Pacific Northwest!
Flippers4pups
Fins up since 1993
View BadgesExcellence Award
Reef Tank 365
Article Contributor
Reef Squad Emeritus
Hospitality Award
Rumor is that I have the most biodiverse rock in the Pacific Northwest!
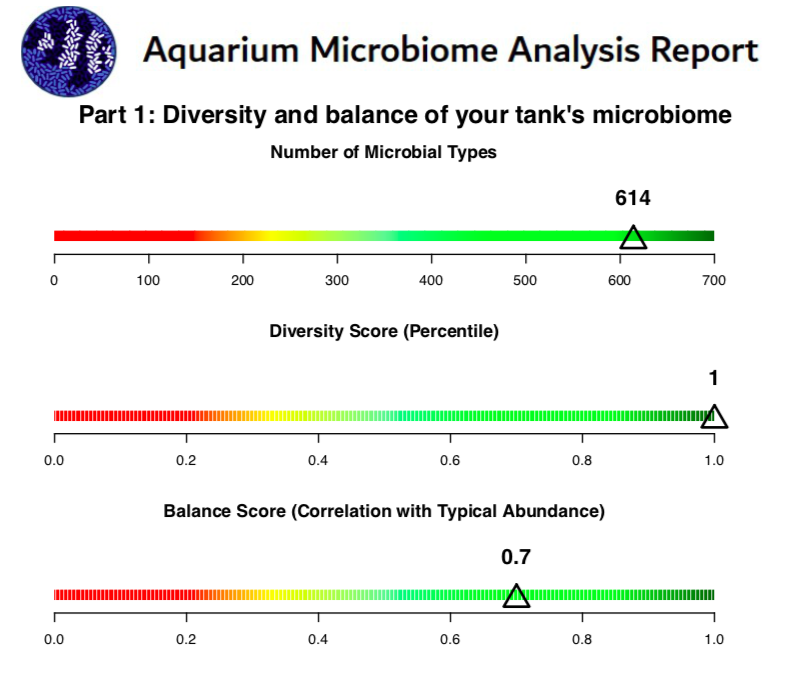
We are going to see more of this in the future!
So are we saying that we should all share little vials of tank water with each other, to share microbes and have maximum variation?
I could see a little "Vial swap" stand setup at frag swaps lol
Vials marked from all the most diverse reefs around the world even could be a possibility.
That’s a fine assertion.
Thank you, Randy.
How do you know it is true?
"Not true" was referring to the sampling protocol. I have one of the kits here in my lap, and I can tell you definitively that it samples both biofilm and the tank water.
As for whether the water samples will miss some bacterial species, my evidence is more speculative. As anyone who has cycled a sterile tank can report, surface-living bacteria don't just stay in place. All tank surfaces are in contact with water, so those microbes should be represented in the water sample to some degree. What I can say from the Aquabiomics report on my tanks, is that it shows a lot more than just planktonic bacteria.
But Eli is the one who has been in the marine microbial ecology literature for the past few years as he developed his sampling protocol. He's 20 miles down the road from me so I have been watching this project develop with much interest! As he said in his post today, he will be presenting evidence supporting his sampling protocols soon.
Different environments will also have different species. The surface of a pipe will be very different than down in sand, on an organism like macroalgae, in light vs dark, etc.
As for sampling the biofilm of the return pipe, versus other surfaces, sampling this surface was ideal because it is the one surface besides glass that pretty much every tank shares.
This is the most interesting and fascinating subject of our hobby to me. Truly awesome.
Can't wait to learn and have more philosophical discussions as this subject progresses.
I personally have a slight obsession with bacteria / "life" in regards to aquariums and the relationship of our ecosystems, the Food chain on the microbial level.
& welcome Eli
Can't wait to learn and have more philosophical discussions as this subject progresses.
I personally have a slight obsession with bacteria / "life" in regards to aquariums and the relationship of our ecosystems, the Food chain on the microbial level.
& welcome Eli
Last edited:
- Joined
- Mar 2, 2019
- Messages
- 893
- Reaction score
- 1,261
One aspect of this that would be very interesting is comparing the microbiome from a "normal" (whatever that means!) reef tank to the microbiome from a carbon-dosed tank. Many of us have noticed that one can go through a few weeks of carbon dosing on a tank with relatively high nitrates and phosphates, have those levels drop significantly, then discontinue carbon dosing and have the NO3/PO4 levels stay low, despite heavy feeding. It'd be interesting to know if carbon dosing is changing the overall microbial mix in the tank so that nitrate reduction occurs regardless of whether ethanol/vinegar/sugar is continuously added.
- Joined
- Sep 8, 2019
- Messages
- 2,303
- Reaction score
- 2,859
A very interesting first step into a new frontier.
It is going to take several years before we gather enough data so that proper actions can be made on the results, but making that database is one of the great thing about this hobby.
It is going to take several years before we gather enough data so that proper actions can be made on the results, but making that database is one of the great thing about this hobby.
Randy Holmes-Farley
Reef Chemist
View Badges
Staff member
Super Moderator
Excellence Award
Expert Contributor
Article Contributor
R2R Research
My Tank Thread
- Joined
- Sep 5, 2014
- Messages
- 67,160
- Reaction score
- 63,517
What about DNA from the bacteria on the surfaces getting into the water column? Does the analysis somehow only pickup DNA from suspended organisms?
I have no idea what levels they can detect and what exactly that translates to. I can’t see how one could quantify the number of benthic bacteria from bits of dna that might be in the water.
- Joined
- Oct 15, 2019
- Messages
- 383
- Reaction score
- 1,600
Agreed, I was interested (and only a little afraid) to look at these groups too.One thing that will be interesting is to see the relative abundance of vibrio and mycobacterial DNA in the samples; two groups that can potentially infect the aquarist.
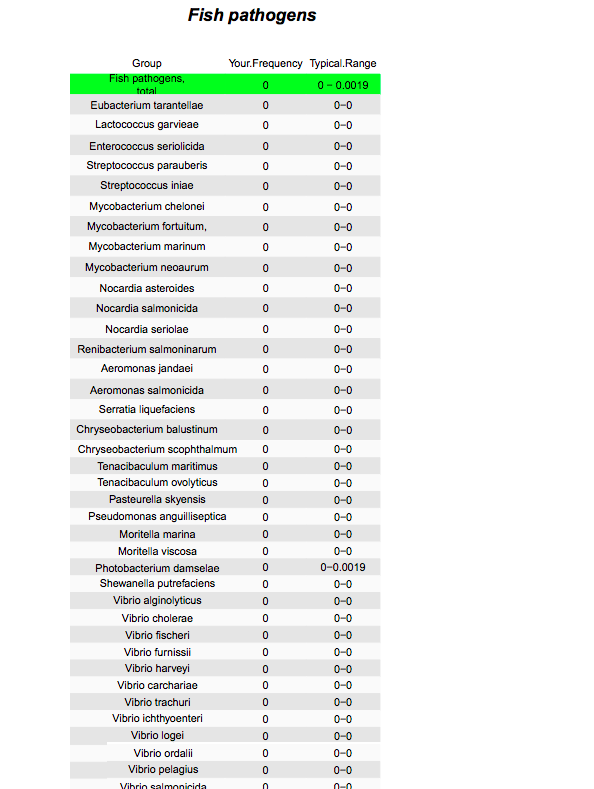
I found bacteria from the family Vibrionaceae in every tank, at reasonably high levels in most tanks and very high levels in a few (average = 4.3% of the community; range = 0.8 - 11.8%).Most of these were in the genus Vibrio (average = 2.8% of the community; range = 0.7 - 11.1%).
This genus includes lots of pathogens of fish, corals, and even humans. Happily, I found no evidence of known coral or human pathogens including Vibrio cholerae, the pathogen responsible for cholera.
Similarly, most samples contained bacteria in the genus Mycobacterium (average = 1.6% of the community; range = 0 - 15.5%). But I found no evidence of any known human pathogens in these tanks, including Mycobacterium marinum, which can cause infections in aquarium hobbyists.
I want to put two important disclaimers here for these statements.
1. I make no claims about human health or safety, and can only refer any such questions to a trained medical professional. My service is intended for hobby, research, and educational purposes only.
2. (More interestingly) What does absence mean? I analyzed about 10,000 DNA sequences per sample. Some microbes present at low levels were detected only once. Very rare microbes present at lower levels would not be detected. 10,000 sequences provides a very thorough sampling of the community, but no realistic sampling of the community can be truly complete.
In other words, like any quantitative analysis, there is a limit of detection in testing microbial communities.
So we should not say these pathogens were absent, but rather that we did not detect them, given this level of sequencing.
- Joined
- Oct 15, 2019
- Messages
- 383
- Reaction score
- 1,600
Hi Randy,I have no idea what levels they can detect and what exactly that translates to. I can’t see how one could quantify the number of benthic bacteria from bits of dna that might be in the water.
Thanks for raising these points, it's always good to have the perspective of someone trained to think quantitatively. The point about surface-associated versus water column bacteria is an important one. In fact, as I think you suggested, in the marine environment we find that there are different communities associated with each microhabitat.
And several of you are rightly wondering about this because many of the microbes that process wastes (ammonia-oxidizing bacteria & archaea [AOA & AOB] and nitrite-oxidizing bacteria [NOB]) are primarily associated with surfaces.
In fact, in the tight confines of a recirculating aquarium, these surface associated microbes are also shed into the water column. In addition to the hobbyist tanks, I also sampled the sand and water in one of my experimental tanks. A few key findings:
1. All the same groups of AOA, AOB, and NOB that I found in sand were also found in my survey of water in hobbyists' tanks.
2. As expected, these groups are much more abundant in surface-associated communities than in the water. For example, Nitrosococcus was 2.8% of the sand community but only 0.06% of the water community (more than 40-fold less abundant than in the sand). Other groups present at lower levels in sand were below the limit of detection in water. For example, the AOA with a tongue-twiser of a name (Cenarchaeaceae) made up 0.05% of the sand community but were not detected in analysis of the same tank's water.
As a result, testing the water alone was not sensitive enough. Several of the mature reef tanks sampled in this study lacked evidence of NOB, despite having effective biological filters capable of producing NO3.
To address this, current kits include an additional step to sample the biofilm. Since many people don't have sand, I chose to sample biolfilms in pipes. These are known to house the groups of microbes we're interested in, based on studies of water treatment plants and recirculating aquaculture systems. So all current and future samples will include both fractions.
-Eli
- Joined
- Sep 8, 2019
- Messages
- 2,303
- Reaction score
- 2,859
I have no idea what levels they can detect and what exactly that translates to. I can’t see how one could quantify the number of benthic bacteria from bits of dna that might be in the water.
Yeah that thought crossed my mind also. I figured that he tested something like 1ul samples from the water with maybe ten different syringes spread into the sample to pull it all out at the same time. Then he checks to see how many of those 1ul tests have what kinds of DNA. A total guess on my part but unless multiple tests are run on very small simultaneous samples I cannot see how you can get any kind of count.
Similar threads
- Replies
- 2
- Views
- 688
- Replies
- 0
- Views
- 82
- Replies
- 9
- Views
- 172
New Posts
-
Help with a fixture for ReeFi Uno overhead mount…tank is 59” wide
- Latest: afternoondelight
-
-
-
-

















